Principle of Simple Sequence Repeats (SSR) and Their Application in Plant Breeding
Since life began on earth about 3 billion years ago, the biodiversity of the plants on our planet have gone through numerous natural selection and evolutionary changes, producing a rich and diverse ecosystem supporting life in various geographical regions and climates. In addition, when human race have arisen as a superior species, various plant species were domesticated into crops to produce various valuable resources. For example, wheat and pea had been domesticated as early as the Neolithic Ages, forming the basis of agriculture in the Middle East, North Africa, India, Persia and Europe. Since crops like rice, wheat, corn, pea are important staple food, while cotton gives textile and clothing, these crops are very important for food security and economy via trading. Hence various efforts to improve these crops have been done since the ancient days via crossbreeding and selective breeding, yielding improved versions of the crops we know of today.
Get Help With Your Essay
If you need assistance with writing your essay, our professional essay writing service is here to help!
However, it was not until the 17th century that the term taxonomy has been introduced by Carl Linnaeus, which systematically categorizes and document living organisms scientifically. Hence, the ancestry and roots of the crops we are planting today are not clearly characterized and remains shrouded in mystery. Still, classical plant breeding practices still progressed, which involves selection of desired traits based on the observable phenotypical characteristics, or morphological markers. This method of selective breeding, however, has some flaw and weaknesses. This is because molecular markers are often masked by environmental factors, or are presented only during late stages of growth, and are sometimes interpreted differently by different individuals. Today, breeding of crops are facilitated by more systematic marker assisted selection methods such as RAPD, RFLP, AFLP, SNP, SSR and ISSR. These methods exploit inherent molecular markers present in the genome of plants to give comparable, consistent, DNA-based polymorphic patterns unaffected by environmental or other external factors.
Simple Sequence Repeats (SSR), also known as short tandem repeats (STR), or microsatellites, is one of the molecular markers commonly used today. As a type of variable number tandem repeat, its principle revolves around repeats of sets of dinucleotide, trinucleotide or sometimes tetranucleotide repeats. The polymorphism itself arises due to the different number of these repeating nucleotide sets, ranging from 3 to 100 times depending on the species. In plants for instance, AT dinucleotide repeats are very common. The AT dinucleotide will occur in alleles, and the number of AT microsatellite repeats can differ between the alleles. These short repeating regions typically occur in the non-coding regions of the genome known as the introns. Hence, they usually do not cause mutations or diseases. However, there are trinucleotide SSRs that are located in the coding exons, they can exist because they do not alter the reading frame of the sequence. In fact, according to Marcotte (1998), 20% to 40% of the proteins in mammals contain repeating sequences of amino acids caused by SSRs. In rare cases, SSRs in the coding region also have been known to cause diseases. For example, Huntington’s disease occurs when CAG trinucleotide repeats span beyond normal copy number. SSRs can also be categorized into 3 types besed on the manner they are arranged, namely simple, compound, and interrupted SSRs. As the name implies, simple SSRs are simply an uninterrupted segment comprising one type of repeating motif, whereas compound SSRs involve 2 or more different types of repeating motifs. Lastly, interrupted SSRs have random nucleotides sandwiched between the repeating units, interrupting the SSRs.
 Figure 1. An illustration of microsatellites of AT dinucleotides. The polymorphism occurs when the upper sequence has 7 repeats of the dinucleotide while the lower strand has 9 repeats.
Figure 1. An illustration of microsatellites of AT dinucleotides. The polymorphism occurs when the upper sequence has 7 repeats of the dinucleotide while the lower strand has 9 repeats.

Figure 2. An illustration describing how simple, compound, and interrupted SSRs are arranged.
The discovery of SSRs actually dates back to the 1960s. It came about when simple repeats scattered in eukaryotic genomes were identified through density gradient centrifugations of randomly sheared genomic DNA. Satellite peaks were found in these sheared DNA, and sequencing revealed repeating motifs of variable length in these sequences, ranging from a single base to several thousand bases. Later, satellites with 10 to 30 repeating motifs were isolated, and these were called minisatellites. It was until 1982, when Hamada and his colleagues demonstrated the existence of dinucleotide poly-CA repeats in eukaryotic genomes, only then the term microsatellites was coined. Since the repeating motifs are usually just one to four bases long, they are then simply called simple sequence repeats, SSR. Eventually, it was demonstrated that SSR polymorphisms could be easily detected by using PCR with 2 primers flanking the SSR region. This discovery in turn led to the development of numerous SSRs in various species and their association to link various loci in chromosomes. The earliest evidence of the occurrence of SSRs in plants was shown when phage libraries of tropical tree genomes was screened via hybridization with poly G-T and poly A-G oligonucleotide probes. This had shown that SSR is also vastly abundant throughout a wide array of planr genomes. Since then, SSRs have been adopted as a popular molecular marker choice for plant genetic analysis and population studies.
It is still not certain how SSRs came about. However, there are several early theories that attempt to explain the polymorphism. It was initially thought that the difference in length of SSRs was caused by unequal crossing over between the repeating units during meiosis. The variation of the SSR bases on the other hand, is hypothesized to be due to a process called DNA replication slippage. Both these mechanisms have the ability to either add or remove the number of repeating units in DNA sequences, causing varied number of repeating units ranging from 2 to 40. Generally, SSRs that have longer repeats will be more polymorphic that others that are shorter in length. It has also been reported that different species will usually preferentially harbor different SSR motifs. The reason for this however is still a mystery.
 Figure 3. Diagram of the mechanism of Replication Slippage during DNA replication.
Figure 3. Diagram of the mechanism of Replication Slippage during DNA replication.
 Figure 4. Diagram demonstrating unequal crossing over during meiosis.
Figure 4. Diagram demonstrating unequal crossing over during meiosis.
The analysis of SSRs involves amplifications using polymerase chain reactions (PCR). A pair of primers flanking the microsatellite region will be used for the PCR. The primers will anneal to each side of the SSR region, and will eventually yield high quantity of PCR product comprising of the repeated SSR sequence. Recall that the number of repeating units will vary between different species, hence the length of these PCR products will also be different. There are 2 commonly used methods to analyze the PCR products. One of which involves gel electrophoresis using agarose or polyacrylamide gels. Because the PCR products are of varying length, they will have different molecular weight, and will migrate at different rates across the gel during electrophoresis. Hence, they will eventually separate on the gel, longer sequences will migrate slower and will remain at upper regions of the gel whereas sequence with shorter repeats will migrate faster towards the lower molecular weight regions of the gel. This therefore yields a visible banding pattern, allowing the experimenter to distinguish the samples based on the bands they yield. For gel electrophoresis, using polyacrylamide gels instead of agarose gels will provide better separation, allowing visualization of bands differing as little as a single base pair. This is particularly useful in resolving SSR polymorphisms with very minute difference in base pairs. The drawback however is, polyacrylamide is toxic and very expensive. Thus, an alternative is to use agarose gels of higher concentration. Another method that can be used is capillary electrophoresis. The mechanism of separation is somewhat similar compared to that of gel electrophoresis, except that it is done in a narrow capillary tube. The PCR product migrating through the capillaries will be excited via exposure to UV light, and a detector will detect the signals produced by the DNA. The signals will then be displayed as a graph, with peaks indicating the detection of DNA. The time whereby each peak is detected will correlate to the size of the fragment. The smaller the fragment, the faster it migrates through the capillary, and the earlier it will be detected.
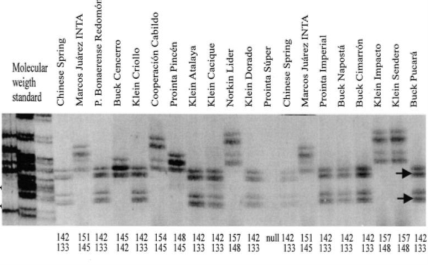 Figure 5. Gel photo depicts an example of SSR-PCR results of 20 different wheat cultivars.
Figure 5. Gel photo depicts an example of SSR-PCR results of 20 different wheat cultivars.
 Figure 6. An example of signals detected on a gene locus in Arabian Oryx using capillary electrophoresis.
Figure 6. An example of signals detected on a gene locus in Arabian Oryx using capillary electrophoresis.
Besides of the 2 PCR based SSR method mentioned before, there is also another PCR amplification based analytical method that also involves simple sequence repeats. It is called inter-simple sequence repeats (ISSR). ISSR analysis is slightly different from SSR PCR, the main difference involves the primers themselves. ISSR primers are short oligonucleotide sequences that are complementary to the repeating SSR sequences. The binding of the primer at these regions will in turn amplify the sequence sandwiched between the SSR repeats. The PCR products can be then separated in the same way, via gel electrophoresis for visualization. The advantage of ISSR analysis is that no prior sequence knowledge is needed, only oligonucleotides of different repeating motifs are needed. However, the drawback is that molecular nature of the polymorphism remains unknown unless the fragments separated in the gel is extracted and sequenced.
Find Out How UKEssays.com Can Help You!
Our academic experts are ready and waiting to assist with any writing project you may have. From simple essay plans, through to full dissertations, you can guarantee we have a service perfectly matched to your needs.
View our academic writing services
We have now discussed how SSRs can be analyzed via PCR amplification using flanking primers. So how were these primers designed? One of the approaches involves searching genome databases that are available using various bioinformatic tools. There are many fully characterized genes, fully sequenced cDNAs available on online databases such as PubMed, GenBank and EMBL. A search for SSR sequences can be performed on this wealth of information, and then flanking sequences of the SSR regions can then be used to design the desired primers. Moreover, there are now even custom-made web based SSR search softwares designed for this purpose alone, such as MISA (MIcroSatellite), CUGssr, SSRSEARCH, and Sputnik. However, this computational search approach has its limitations. When we need to do SSR analysis in some minor or less known
Cite This Work
To export a reference to this article please select a referencing style below:


