A High-Throughput Screening Triage Workflow to Authenticate a Novel Series of PFKFB3 Inhibitors: Paper Summary
A high throughput screening was carried out of human 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), which resulted in multiple compound series with the potential for further development[1]. Multiple screening methods were carried out, including a Primary PFKFB3 HTS spot-test assay with an IC50 follow up, an Orthogonal IC50 assay and Enzyme Ratio Testing; these methods shall be outlined throughout this summary.
Primary PFKFB3 HTS spot-test assay + IC50 follow up
The screening of compounds by St-Gallay et al was performed using an ADP-Glo format [2] – a dual-step process. The protein of interest (PFKFB3) was incubated with two compounds, fructose-6-phosphate (F6P) and ATP in a buffer solution, containing 5 mM MgCl2, 5 mM KPi at PH7, 200 mM KCl, 0.1 mM Triton X-100, 50 mM HEPES and 1mM dithiothreitol (DTT); thus giving the terminal concentrations of PFKFB3, ATP and F6P. (8.75 nM, 34 µM and 15 µM respectively, as outlined below in Figure 1.)
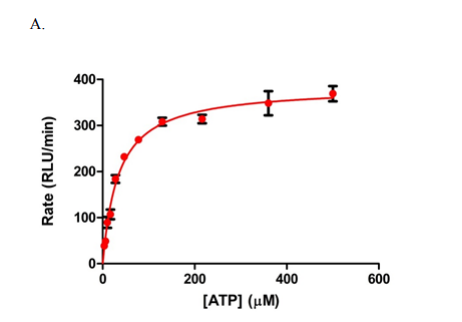
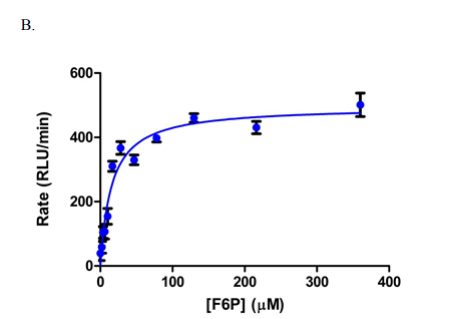
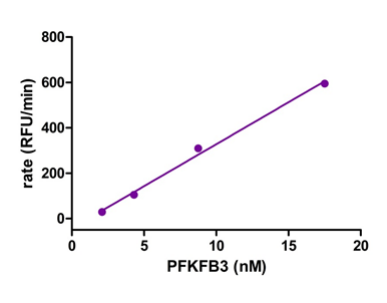
Figure 1: Graphs outlining the final concentrations and rate of luminescence (RFU/RLU per minute) of ATP, F6P and PFKFB3[1]
The primary run was undergone in the 1536 well format, whereby 1µL of enzyme and 1µL substrate additions were made simultaneously, to give the final volume of 2µL per well. All other assays were prepared using a very similar method, by addition of a fraction of solution containing enzyme (reactions already initiated) with an equal volume of substrate solution. After approximately 60-70 minutes of time passing, ADP-Glo reagent was introduced to the wells in order to terminate the kinase reaction, and exhaust any remaining ATP.
Get Help With Your Essay
If you need assistance with writing your essay, our professional essay writing service is here to help!
After approximately another 40 minutes, a kinase detection reagent was added, in order to convert adenosine diphosphate (ADP) back to its original ATP form. The newly formed ATP could then be quantitated by the luciferin/luciferase reaction, where for each molecule of ADP that was produced, it was assumed that 1 molecule of ATP had regenerated. Luminescence could then be determined and used as an indicator of enzyme inhibition; it was found that molecules that did indeed inhibit PFKFB3 reduced the luminescent signal when compared to uninhibited enzyme.
Both the primary screen and follow up followed the same terminal conditions, in both a 384 well (low volume plate) and 1536 well format. Both substrate and enzyme additions were added at a 1:1 ratio, although the total volume was 2µL per well for the 1536 format and 5µL per well for the 384 well format. The ADP-Glo was added stoichiometrically (1:1:2 ratio as recommended by the manufacturer) in regards to volume/ADP-Glo reagent/kinase detection reagent [1].
Bovine Serum Albumin (BSA) was considered for the determination of non-specific binding, however as it is well known that BSA has a tendency to bind drug-like molecules[1], of which many are confirmed to inhibit enzymes, so there was a real risk of removing genuine hits through false negative results. Therefore it was decided that the protein of interest (PFKFB3) was to be used, with the added benefit of the ability to construct a kinetically equivalent assay at both of the concentrations outlined above.
Full IC50 (follow up) curves were produced for a specific subset of hit compounds, these were identified from the initial library of 837387 compounds; A comparson of these results with the orthogonal inhibition assay can be found in Figure 2 below. Finally those selected compounds were tested within an artifact IC50 format, whereby the detection part of the ADP-Glo assay was run again, this time without PFKFB3 and instead with an equivalent concentration of ADP/ATP – which was a substitute for what would have been produced by the aformentioned kinase reaction. Therefore if any of the selected compounds interfered with the detection of the mixture, there would be a notable decrease in signal observed; however none of compounds tested were found to cause this effect.
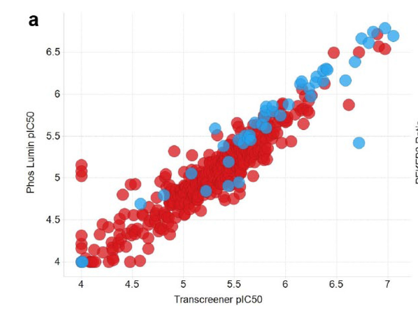
Figure 2: A graph outlining PFKFB3 pIC50 when comparing luminescence (ADP-Glo assay) versus the Transcreener technology(orthogonal inhibition assay) [1]
Orthogonal inhibition assay
The compounds brought forward for an IC50 follow up using the ADP-Glo assay after the initial HTS were also tested used a Transcreener ADP Fluorescent Intensity Assay [1]. Due to the fact that this assay is based on detecting ADP, any class of compounds /enzyme that produces ADP is compatible with Transcreener technology [3]. The process of assay preparation was designed to match the conditions used in the previous ADP-Glo assay, however it was found that instead of a protein concentration of 8nM being left for 60 minutes, a concentration of 4nM being left for 120 minutes gave a linear reaction and more appropriate signal window, as outlined in Figure 3 below.
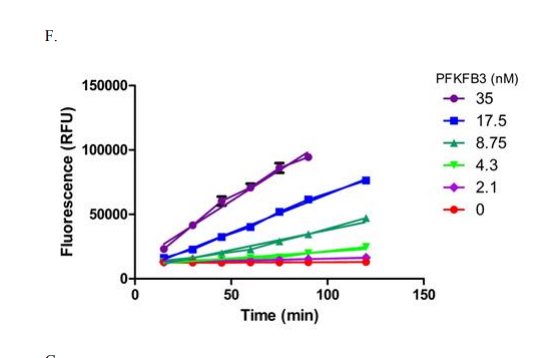
Figure 3: Using ADP-Glo conditions, linearity of a reaction time course was measured with various concentrations of PFKFB3 for the orthogonal inhibition assay [1]
The assay was again performed in two steps, first PFKFB3 was incubated with the compound, ATP and F6P, in the same buffer solution used prior; the only difference in final concentration was PFKFB3 at 4.3nM instead of 8.75nM, again highlighted by Figure 3, and the solutions were left for 2h instead of 1h.
ADP was able to be detected due to a tracer (ADP Alexa 594) that was bound to ADP2 monoclonal antibody [1], as the tracer is displaced by ADP it is released into solution and causes a positive increase in fluorescent readout. Consequently, there is a correlation between kinase activity and an increase in fluorescence due to a rise in ADP production.
Find Out How UKEssays.com Can Help You!
Our academic experts are ready and waiting to assist with any writing project you may have. From simple essay plans, through to full dissertations, you can guarantee we have a service perfectly matched to your needs.
View our academic writing services
Using a black, low volume 384 well plate, after 2 hours of incubation 5µL (2.5 µL of PFKFB3 and 2.5 µL of ATP/F6P) of solution was added to each well. After approximately a further 60 minutes, fluorescence readings of the plates were taken. The IC50 values were then generated from these results, and compared against the results achieved through the ADP-Glo assay, as again can be seen in Figure 2. A positive correlation was observed between the two assays in regards to compound potency values [1] suggesting a high level of accuracy in IC50 readings.
Enzyme Ratio Testing
A secondary enzyme test occurred in order to determine nonspecific compounds effects, thereby allowing separation of potential hits that are time-dependant or enzyme-concentration dependant. By increasing the PFKFB3 concentration 10-fold, and simultaneously decreasing the incubation time by the same amount (60 minutes decreasing to 6 minutes), it was possible to ascertain effects (both time and concentration dependant) that potentially occurred due to the compound behaviour within the enzyme assay, for example compound aggregation. [4]
ERT was an excellent secondary filter that was used to specify more promising candidates from the initial hits through both HTS and the following assays.
Conclusion
St-Gallay et al were able to benefit from the highly accurate and selective HTS and following assays in order to determine ‘right’ compounds with positive lead-like properties [5]. By taking advantage of assays designed around ADP production and the detection of fluorescent tags, with powerful filtering and analytical techniques (Enzyme Ratio Testing, Isothermal Calorimetry and X-Ray Crystallography) the dihydropyrrolopyrimidinone series of compounds [1] were identified as a potential option in developing potential treatments for tumours – by targeting the increased glycolytic flux that occurs in tumour growth [6].
References
- [1] Stephen A. St-Gallay, Neil Bennett, Susan E. Critchlow. A High-Throughput Screening Triage Workflow to Authenticate a Novel Series of PFKFB3 Inhibitors, Sage Journals. SLAS Discovery. 2017, Vol. 23, 11–22.
- [2] Zegzouti, H., Zdanovskaia, M., Hsiao, K.. ADP-Glo: A Bioluminescent and Homogeneous ADP Monitoring Assay for Kinases. Assay Drug Dev. Technol. 2010, 7, 560–572.
- [3] Kleman-Leyer, K. M., Klink, T. A., Kopp, A. L.. Characterization and Optimization of a Red-Shifted Fluorescence Polarization ADP Detection Assay. Assay Drug Dev. Technol. 2009, 7, 56–67.
- [4] Habig, M., Blechschmidt, A., Dressler, S.. Efficient Elimination of Nonstoichiometric Enzyme Inhibitors from HTS Hit Lists. J. Biomol. Screen. 2009, 14, 679–689.
- [5] Teague, S. J., Davis, A. M., Leeson, P. D.. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed. 1999, 38, 3743–3748
- [6] Buerkle, A., Weber, W. Imaging of Tumor Glucose Utilization with Positron Emission Tomography. Cancer Metastasis Rev. 2008, 27, 545–554.
Cite This Work
To export a reference to this article please select a referencing style below:


